Machine learning (ML) is a powerful tool for science. It can automate expensive processes, reveal subtle patterns in data, and even guide experimental design. But in practice, building ML systems is slow, complex, and highly manual. Every dataset requires custom preprocessing. Models must be hand-picked, tuned, and validated. Implementing interpretability – essential for scientific trust – adds another layer of complexity. It often takes weeks or months to yield insight.
Discovery Engine changes this.
It’s a general-purpose system that automates the entire scientific modelling workflow – from raw data to interpretable results. It cleans and processes data, selects and trains appropriate models, extracts robust patterns, and generates structured scientific outputs – all without human intervention.
To validate its capabilities, we set a high bar: replicate five peer-reviewed scientific studies, each of which used machine learning to tackle real-world problems in medicine, materials science, social science, environmental science, and public health. These studies represent the current standard of AI-for-science practice: manually curated datasets, expert-tuned models, and interpretability analyses that took weeks or months to produce.
The Discovery Engine replicated them all, automatically, in a matter of hours.
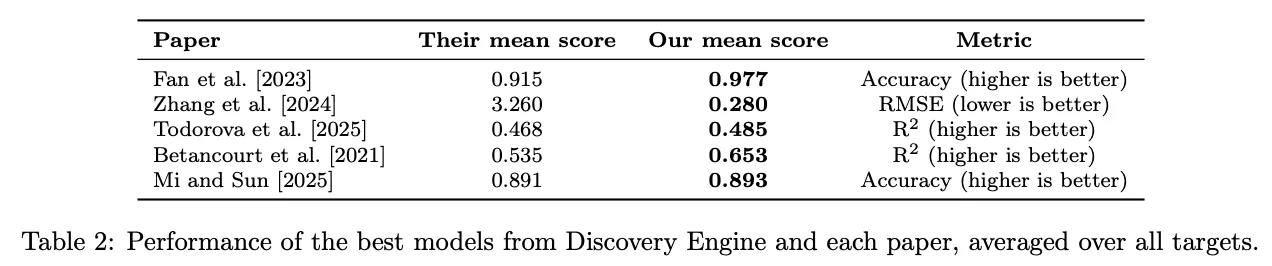
In every case, it matched or exceeded the predictive performance of the original study. It confirmed key scientific findings through interpretable model artefacts. And in several cases, it uncovered new, domain-relevant insights that had been missed the first time around.
Hepatitis C Diagnosis
Original study: Fan et al., BMC bioinformatics, 2023.
The authors trained several classifiers to predict hepatitis C infection from blood markers, selecting a Bayesian-optimised random forest as the most performant. They used SHAP and LIME to identify the most important features learned by the model, which included AST, GGT, and ALP.
The Discovery Engine model outperformed the original on all metrics except AUC, on which it came close. It confirmed the same key biomarkers as being most important, but also found a nuanced interaction: ALP values that typically suggest low risk become high-risk when paired with elevated AST. This was missed in the original paper – understandably, as this kind of nonlinear, combinatorial insight is extremely hard to find with standard tools.
Concrete Strength Prediction
Original study: Zhang et al., Nature Scientific Reports, 2024.
This Nature paper evaluated four model families with hyperparameters tuned via random search, to predict concrete compressive strength from its various components. Their best-performing model was an ensemble of boosted decision trees (LightGBM). They also used SHAP to identify the most influential variables, which were age, water/cement ratio, slag content, and water.
Discovery Engine’s neural network reduced prediction error by an order of magnitude, achieving an RMSE of 0.28 (vs 3.26). It actually identified different features as most predictive – but since it has learnt to represent relationships not captured by the original model (evidenced by its superior performance) this is unsurprising. One pattern found was that low slag content combined with certain aggregate/cement ratios dramatically reduced strength, echoing known materials science principles, but not captured by the original model.
Climate Beliefs and Behaviour
Original study: Todorova et al., npj Climate Action, 2025.
The study used gradient-boosted decision trees to predict a set of climate change related beliefs, based on psychological and socioeconomic predictors like environmentalist identity, trust in science, location, HDI, etc. They also used SHAP for feature importance.
Discovery Engine matched or surpassed the original predictive performance across all four target variables. It agreed on three of the four most important features quoted in the paper, and also extracted novel patterns, including one showing low climate belief among people with low trust in science and high exposure to climate risks – an interaction hinted at in regional studies but never surfaced at scale.
Ozone Pollution Forecasting
Original study: Betancourt et al., Earth System Science Data, 2021.
This paper established a benchmark for modelling tropospheric ozone using global station-level data, aggregating five years of ozone metrics across over 5500 monitoring stations. Linear regression, random forests, and shallow neural networks were used to predict multiple metrics of ozone pollution.
The Discovery Engine surpassed their best-performing model on 14 of 15 targets, and demonstrated particularly improved performance for extreme ozone level prediction, which is both scientifically important and typically challenging to predict well. It also uncovered interpretable drivers of these extremes (e.g. warm climates with high nitrous oxide levels predicted ozone spikes) consistent with mechanistic literature on photochemical ozone formation.
Hearing Loss Risk
Original study: Mi and Sun, Hearing Research, 2025.
This study used NHANES data to identify predictors of hearing loss from demographic, clinical and laboratory data. The authors trained several supervised models, including Random Forests, XGBoost, Support Vector Machines, and fully connected neural networks. Their feature importance analysis revealed statistically significant associations between lead exposure, low vitamin E intake, and an increased risk of hearing loss.
Discovery Engine matched the performance of their best model. It also highlighted similar predictive patterns: low blood lead and high vitamin E levels in middle-aged individuals were strongly associated with reduced hearing loss risk, which is supported by independent literature.
Why This Matters
These studies represent hundreds of hours of human effort: experts selecting models, tuning parameters, interpreting results, and contextualising findings. Replicating that process with a general-purpose system – fully automatically, in hours – is a milestone not just for automation, but for scientific replicability. We match or exceed the predictive performance of models built and trained by experts, while also generating interpretable, domain-relevant, and scientifically valid insights.
Most AI tools used in science today are black boxes, or bespoke workflows requiring close involvement from data scientists. They’re powerful, but fragile and expensive. The Discovery Engine is different. It’s robust across domains, adaptable to new datasets, and consistently produces outputs that are not just accurate, but useful – the kind scientists can trust, build on, and publish.
We’re making ML-for-science accessible to all scientists, whether they have the expertise to train a neural network or not. And because the Discovery Engine explains why a model works, and renders the patterns it has learned interpretable, it opens the door to hypothesis generation, mechanism discovery, and ultimately scientific understanding at massively increased speed and scale.
You can read the full technical preprint here.